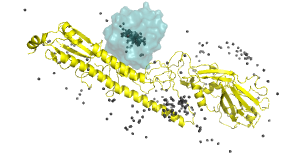
| CAPRI scoreset v2022 |

| Introduction | Browse | Download | Help |

The CAPRI scoreset v2022 is a follow-up to the initial Score_set, published in 2014:
- 2023 The CAPRI scoreset 2022 update (in preparation)
- 2014 Score_set: a CAPRI benchmark for scoring protein complexes
This version is a major update, only partly compatible with the previous version. It contains not only the so-called "Uploader" models already present in the previous set, but also those models submitted by the "Predictor" and "Scorer" groups.
In addition, all files have been homogenized, such that:
- all decoys are superimposed on the receptor entity,
- all chain labeling and residue numbering correspond to the target, and
- all computed assessment quantities are included in the dataset.
The CAPRI scoreset v2022 includes all published CAPRI targets up to CAPRI Round 50, including joint CAPRI/CASP Rounds.
Frequently asked questions
|
Where can I find more information about CAPRI? |
For this we refer to one of the many CAPRI publications:
|
|
|
How big is the database and which files do I - minimally - need to download? |
| The uncompressed size of the database is 57.45 GB, compressed it's 7.24 GB. If you're new to scoring, we recommend you start with the "Scorers" tar file as it represents the smallest set. If on the other hand, you need as many structures as possible (because of clustering, for instance), take the "Uploaders" set. | |
|
How do I use this dataset? |
| You can use this dataset for testing, but also for training. If you use the dataset for both, we do warn against input bias; e.g. if you use 80% of the dataset for training and 20% for testing, there should be no target/interface overlap between the training set and the testing set. Best approach is to use the dataset only for testing, or only for training. | |
|
What is the difference between the P, U and S sets? |
| A CAPRI Round consists of a "Docking" and "Scoring" experiment. For the docking, "Predictors" are asked to submit a set of (up to) 100 models, the first five (or ten) of which are assessed. The full set of 100 models from all Predictors constitutes the Uploader set, from which the Scorers select their set of (up to) ten models to submit. | |
|
How is the difficulty level determined and how many Easy vs Hard targets are there? |
| The difficulty level is provided for every interface of each target is taken from the various CAPRI publications. Since not all publication contain the Medium category, we have here grouped Medium and Difficult together. Of the 148 interfaces, 71 are tagged Easy and 77 Difficult, corresponding to 47.97% and 52.03%. | |
|
Why are there so many incorrect decoys in all sets? |
|
What can I say. Docking is not easy! There are a number of factors that influence the difficulty of any given target. These generally boil down to conformational flexibility and uncertainty. For more information we refer to any one of the CAPRI publications (see first item). |
|
|
Where is the interaction_type annotation coming from? |
| These annotations, "homomeric organization, "enzyme-inhibitor", "artificial binding", et cetera, are manual. | |
|
What is the difference between "bound" and "unbound" docking? |
| For a limited number of - generically difficult to model - targets, the bound conformation of one of the partners was supplied. For these, the docking_type is annotated with "bound". All other targets involved "unbound" docking; an unbound conformation, a template, or only sequence information was supplied. | |
|
What is the difference between the XML and JSON file? Which one should I take? |
| There is no difference. The JSON file is created from the XML file. Use the one that you're most comfortable with processing. | |
|
I don't know how to work with XML or JSON. Do I really need those? |
| No, you don't. All the information about decoy quality can also be found in the CSV file. | |
|
What do all the columns in the CSV file mean? And do I need all of them? |
|
The columns are explained HERE. No, you will not need all of them (but you might). Only the most important ones are included in the XML and JSON files. |
|
|
Why aren't all targets ever presented in CAPRI included in the dataset? |
| Confidentiality is a big thing in CAPRI, as the CAPRI experiment is dependent on experimentalists providing their structure to the assessors prior to its publication. We therefore include only targets with published PDB structures. | |
|
Why do some targets only have Predictor content? |
| Timing in CAPRI is sometimes tight, due to impending publication of the target's associated manuscript. For those targets we were not able to organize a Scoring Round, even though the scoring typically only adds one or at most two weeks to the process. This particularly might happen for the higher impact targets, but it has also happened that an image of the target was published on-line before the end of the Round, leading to the cancellation of the target, or even the entire Round. | |
|
Do the interfaces of a single target together form the assembly? |
| No. It depends on the target. For some targets the interfaces together form an obligate assembly, but for some other targets they may be mutually exclusive. | |
|
Which techniques were used to create the web site? |
| The web site was created using XML, PHP, CSS and JQuery; the images of the proteins using PyMol. | |
|



|